Spot-On analyzes single particle tracking datasets.
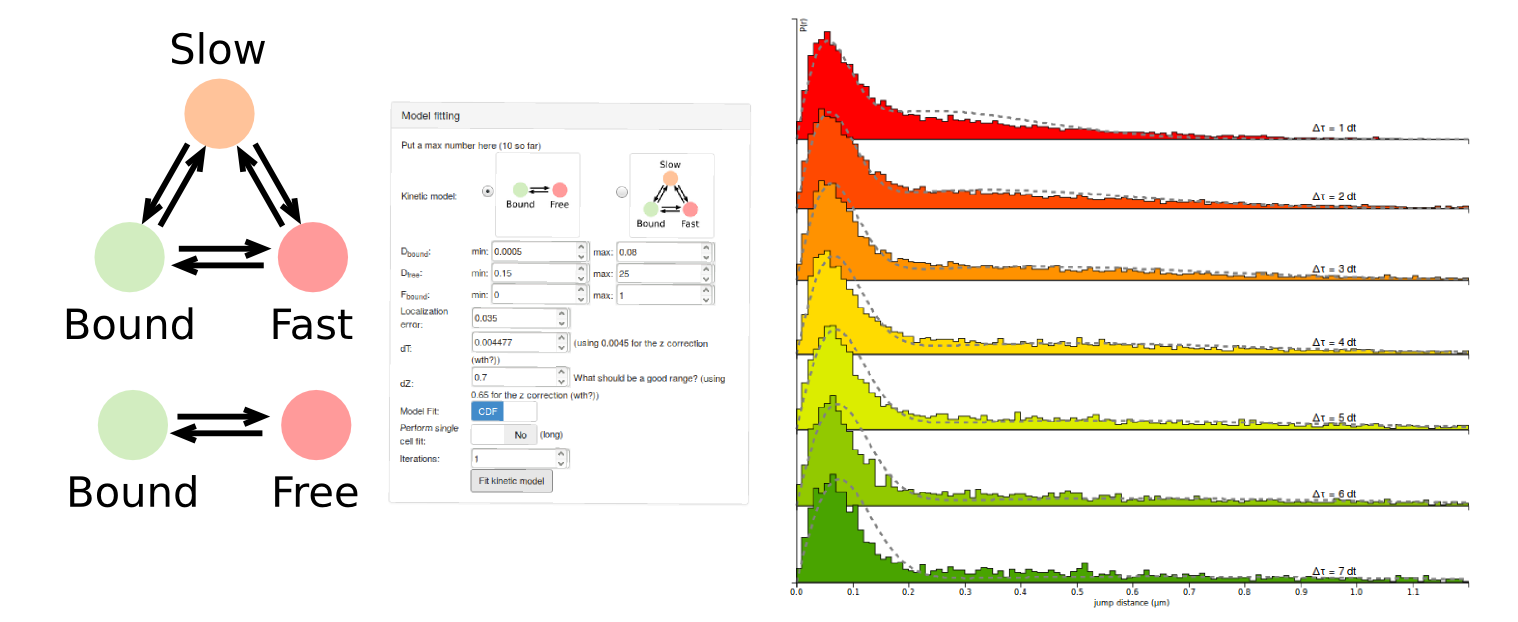
Interactive kinetic modeling
Spot-On comes with a user-friendly interactive interface to perform kinetic modeling of single particle tracking datasets.
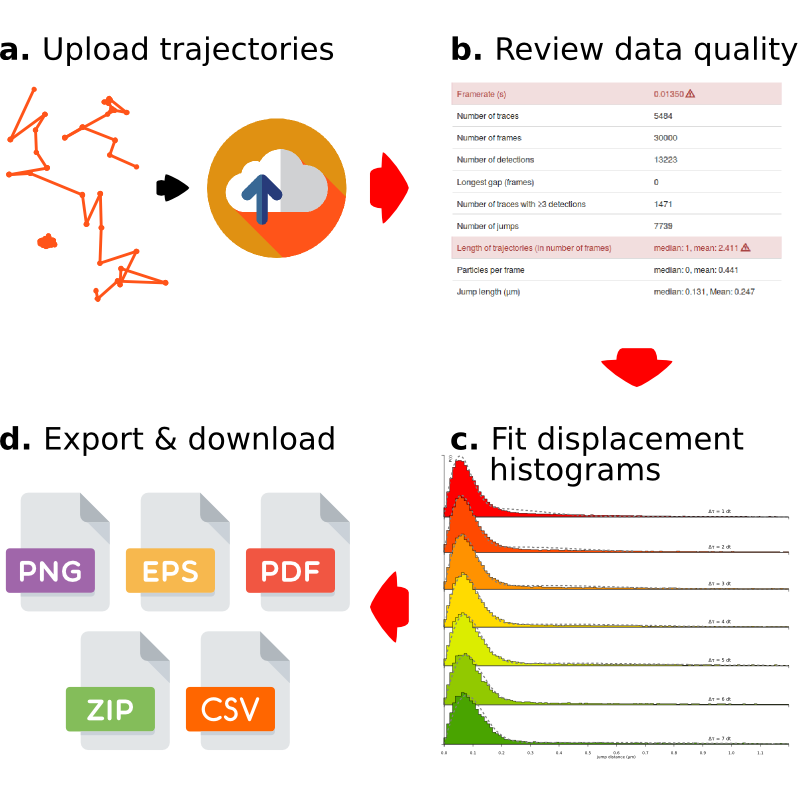
from a wide range of tracking softwares, including a direct upload from Fiji/TrackMate.
Fit a kinetic model to the distribution of jump length. Take into account the finite depth of field.
Save your analyses to a variety of formats (plots, tables, editable files). Ready?
Single particle tracking (SPT) is often seen as less biased and model-dependent than indirect and bulk techniques such as Fluorescence Recovery After Photobleaching (FRAP) or Fluorescence Correlation Spectroscopy (FCS).
Nevertheless, SPT data is subject to a number of biases and uncertainties, which must be corrected for in order to obtain accurate estimates of subpopulations. Spot-On offers a set of guidelines to perform meaningful SPT experiments and provides a comprehensive, bias-aware, framework to analyze the resulting data.

Spot-On fits a two-states or a three-states model to the empirical distribution of jump lengths obtained from a single particle tracking dataset. To do so, Spot-On takes into account a finite detection volume and biases against fast-moving particles. From that, Spot-On extracts kinetic parameters: diffusion coefficients, fraction bound.
Learn more!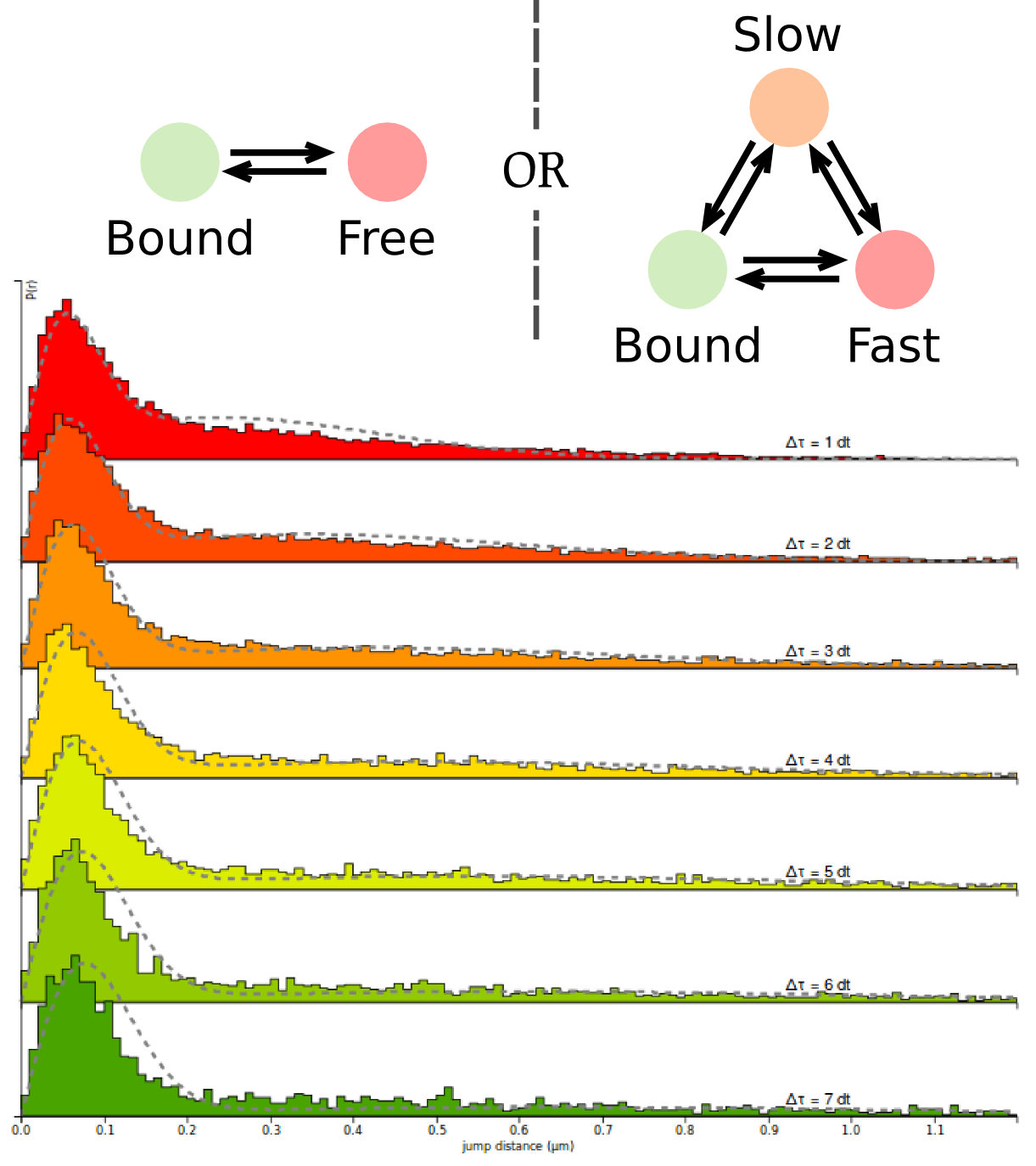
To use Spot-On is straightforward, and Spot-On is fully documented. We have a step-by-step tutorial that details how to use Spot-On from scratch. In particular, the tutorial will guide you through:
Please acknowledge Spot-On in your publications:
Robust model-based analysis of single-particle tracking experiments with Spot-On.
Hansen, A.S.*, Woringer, M.*, Grimm, J.B., Lavis, L.D., Tjian, R., and Darzacq, X.